Our fundamental interest in chromatin is understanding the structural and dynamical features of this assembly that govern genomic regulation and could yield unique therapeutic possibilities. The basic repeating units of chromatin are nucleosomes, which consist of a nucleosome core component (in isolation, this is the nucleosome core particle [NCP]) and linker DNA (typically 10 to 80 base pairs in length).
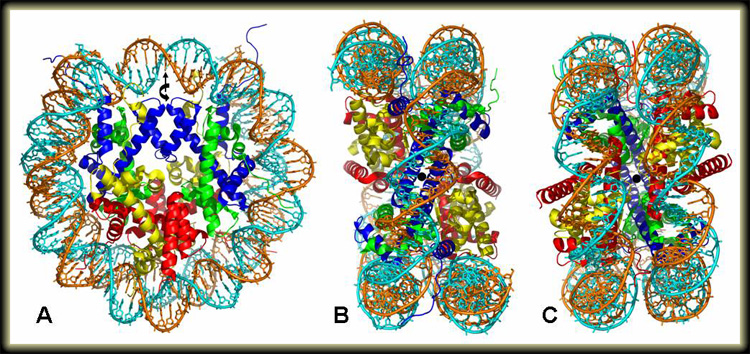
Side (A), top (B), and bottom (C) views of the 1.9 Å resolution crystal structure of the nucleosome core particle (Davey et al., 2002). The two strands of the 147 base pair DNA and the H3 (blue), H4 (green), H2A (yellow), and H2B (red) core histone proteins are colored individually. The view in A is along the DNA superhelical axis, with the pseudo two-fold axis of the particle running vertically through the central DNA base pair (straight arrow). In B and C, the view is along the pseudo two-fold axis, shown as a black dot.
Crystals of the nucleosome core particle
We have analyzed nucleosome structure and stability with many different DNA fragments to derive a general mechanical model that helps explain and predict the sequence-dependent properties of this system. This sheds light on biological questions, but also gives us a variety of substrates to use in our drug development assays and the ability to design DNA sequences tailored for specific chromatin studies.
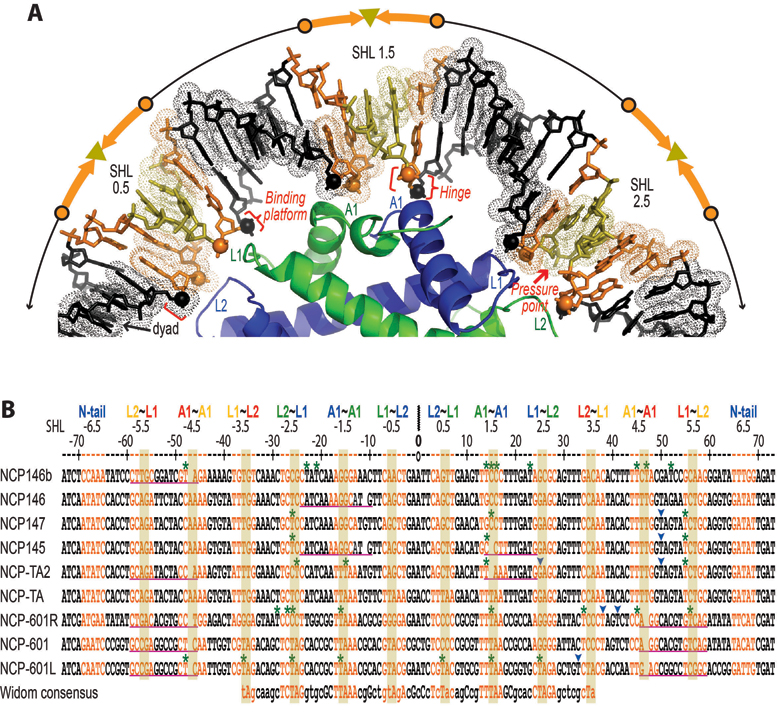
Mechanical scheme for DNA sequence-dependent behavior in the nucleosome (Chua, Vasudevan, Davey, Wu, Davey, 2012)